Description
Most complex traits, such as blood protein levels and memory performance, have a genetic component and many genes are thought to be involved. In genome-wide association studies, known as GWAS, we test whether millions of genetic variants are associated with a measure of interest, such as blood protein levels. These studies are undertaken within CHeBA, with other national and international studies and in collaboration with international consortia.
Aims
To identify genetic variants associated with a variety of measures, such as blood protein levels and cognitive performance.
Design and Method
Genetic variants were genotyped using genome-wide genotyping arrays in participants from the Sydney Memory and Ageing Study (n~925), the Older Australian Twins Study (n~539) and the Sydney Centenarian Study (n~256). Statistical tests are undertaken to find genetic variants associated with many different age-related outcomes including cognitive tests, white matter hyperintensities, hippocampal volume, other brain structures, and plasma protein levels.
Findings
These studies have identified genetic variants associated with a variety of different measures. For example, the results of a genome-wide association study for the blood protein, apolipoprotein H, are shown in Figure 1 using participants from the Sydney Memory and Ageing Study and the Older Australian Twins Study. These results were replicated in an independent cohort. This work has been published by CHeBA researcher, Dr Mather et al (Scientific Reports, 2016).
Our exciting work continues, seeking to better understand how genetics influences different age-related measures in our cohorts of older adults and in collaboration with national and international studies. This research aims to add to our understanding of the molecular processes underlying these traits. In the future it may assist in identifying those who are at risk of age-related decline and disease and facilitate the development of treatment and preventative strategies.
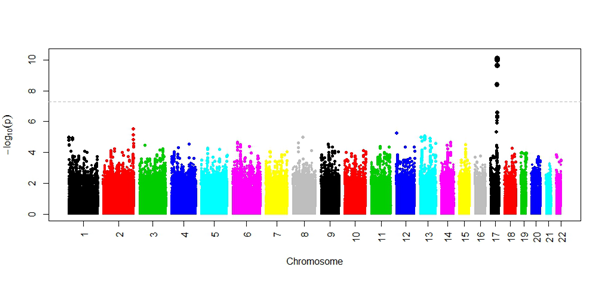
Figure 1: Summary of the genome-wide association study (GWAS) results for blood apolipoprotein H levels. This protein transports lipids in the circulatory system and has been linked to negative health outcomes. Each genetic variant examined is shown by a coloured circle. As shown by the dots above the dashed horizontal line (showing the threshold for genome-wide significance), genetic variants associated with apolipoprote in H levels were found on chromosome 17.
Publications
Published GWAS Studies include:
- Vojinovic, D., et al. (accepted 2018) Genome-wide association study of 23,500 individuals identifies 7 loci associated with brain ventricular volume. Nature Communications
- Davies G., et al. (2018). Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nature Commun, 9, 2098.
- Jiyang, J., et al (accepted 2018) A Meta-analysis of Genome-wide Association Studies of Growth Differentiation Factor 15 Concentration in Blood. Frontiers in Genetics.
- Willems, S.M., et al. (2017) Large-Scale GWAS Identifies Multiple Loci Associated with Hand Grip Strength and Provides New Insights into the Biology of Muscular Fitness. Nature Commun.
- Hibar, D.P., et al., (2017). Novel genetic loci associated with hippocampal volume. Nature Commun., 8:13624.
- Adams, H.H.H., et al. (2016). Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nature Neurosci. 19 (12):1569-1582.
- Ibrahim-Verbaas, C., et al. (2015) GWAS for executive function and processing speed suggests involvement of the CADM2 gene. Molecular Psychiatry, 21(2):189-97.
- Debette, S., et al. (2015). Genome-wide studies of verbal declarative memory in nondemented older people: The Cohorts for Heart and Ageing Research in Genomic Epidemiology Consortium. Biological Psychiatry; 77(8):749-63.
- Davies, G., et al. (2015). Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Molecular Psychiatry, 20(2):183-92.
- Chan, J.P., et al. (2015). Genetics of hand grip strength in mid to late life. Age (Dordr), 37(1):9745.
- Mather, K.A., et al Investigating the genetics of hippocampal volume in older adults without dementia. (2015) PLoS One, 10(1):e0116920.
- Hibar D.P., et al. (2015). Common genetic variants influence human subcortical brain structure. Nature, 520 (7546):224-9.
Project Members:
Project Lead: Dr Karen Mather
Genetics statisticians/bioinformaticians:
Dr Anbupalam Thalamuthu
Dr Nicola Armstrong (Murdoch University)
Dr Rick Tankard (Murdoch University)
Research Assistant: Sri Chandana Kanchibhotla